Technical Tools and Guidance for the Detection and Identification of LMOs
Overview of available detection methods, including validated methods
By: Chris Viljoen, Sarah Agapito-Tenfen, and Gretta Abou-Sleymane
1. General introduction into GM detection methods
A number of methodologies and techniques are available to detect, identify and quantify living modified organisms (LMOs). These methodologies range from those that are fast and more cost-effective, such as lateral flow tests and endpoint PCR, to those that can be more complex, such as quantitative real-time PCR. When planning and setting up a laboratory for the detection and identification of LMOs a choice must be made regarding which methodologies and protocols will be adopted. For service-oriented laboratories, particularly those servicing regulatory authorities, the selection of methods is guided by, amongst other things, the country’s specific regulatory requirements in accordance with national biosafety laws. The methodologies may therefore range from qualitative methods to detect the presence of LMOs, to tests that identify individual LMOs, to quantitative tests that measure the percentage of LMOs present in a sample. Below is a brief overview of some of the more commonly used methodologies for the detection, identification and quantification of LMOs, their strengths and limitations.
2. Introduction to protein versus DNA approaches to GM detection
LMOs are often developed by inserting one or more “genes of interest” which are DNA molecules encoding proteins that confer particular traits of interest, such as insect resistance or herbicide tolerance. Either the DNA or the protein can be targeted for the detection or identification of such LMOs. Both approaches, i.e. protein- or DNA-based methods, have advantages and disadvantages and the adoption of one over the other, or both, will depend largely on the available expertise, infrastructure to handle samples, laboratory equipment and regulatory requirements.
2.1 Protein-based methods for LMO detection
LMO specific proteins (i.e. those produced by the inserted genes) can be detected by antibody recognition of an epitope specific to the transgenic protein. The method of protein testing is either in the form of a lateral flow strip test, a micro-well format as an enzyme-linked immunosorbent analysis (ELISA) or a gel electrophoresis protein immunoblot (also known as western blot)
Protein detection using lateral flow strip tests, ELISA or western blot is performed through a simple procedure of extracting total crude proteins from a sample by adding water or buffer followed by sample homogenization. Protein-based detection methods require the use of antibodies to detect the transgenic protein. Since the process of antibody production is extremely complex and costly, detection using these methods typically relies on the availability of commercial antibodies. For lateral flow strip testing, the strip is placed in the crude protein extract and a positive result is indicated by the appearance of a test line due to the antibody recognition of the transgenic protein. The advantages of this qualitative method are that it is simple to perform, requires little technical expertise or equipment and can be performed at the point of sampling. Electronic devices have also been developed that allow a semi-quantitative interpretation of the result. A disadvantage of this method is that its sensitivity is dependent on the binding affinity between the antibody and the protein.
The ELISA based approach to LMO detection follows the same crude protein extraction step as the lateral flow strip. However, the antibody is used to pre-coat the inside of a micro-well plate. Following a series of steps which allow the target protein to bind to the antibody, the cell debris, including other proteins, are removed from the plate through a series of wash steps. The bound protein is detected through a colour reaction that can be read through visual inspection or by an optical plate reader. This method produces a qualitative result if read through visual inspection. However, a quantitative result can be obtained if the necessary protein standards are included on the plate and an optical plate reader is used to evaluate the intensity of the colour reaction resulting from antibody recognition of the target protein in the LMO. The advantages of ELISA LMO testing are that it can produce a qualitative or quantitative result and is more sensitive than the lateral flow strip method.
For western blotting, the extracted crude proteins are separated according to their size by gel electrophoresis. Following this, the proteins are transferred from the gel to a membrane for detection of the target protein. Usually this step of the process involves two antibodies: firstly a primary antibody that is specific to the target protein followed by a secondary antibody, which is linked to a reporter molecule that binds to the primary antibody. After the excess antibody is removed from the membrane, the secondary antibody is typically visualized by colorimetric, chemiluminescent or fluorescent methods performed by either colouring the membrane itself or exposing it to a light sensitive film, such as x-ray film. Once the membrane or film is developed, the presence of the transgenic protein is indicated as a distinct band on the membrane or film. The advantage of this method is that it is sensitive and may detect different isoforms of the target protein. A disadvantage of this method is that the primary antibody may cross react with native forms of the same protein that may be present in the organism.
2.2 DNA-based methods for LMO detection
DNA-based methods for LMO detection and identification are based mainly on the use of the polymerase chain reaction (PCR). PCR is a method that employs synthetic DNA oligonucleotides, so called “primers”, to replicate or “amplify” targeted regions of an inserted DNA sequence that is present in the LMO. The amplified product can then be detected to determine whether or not DNA originating from an LMO is present in a sample.
DNA-based methods require the extraction of DNA prior to PCR detection. It is important to determine which DNA extraction method is appropriate for a particular LMO. While most extraction methods employ a cetyltrimehylammonium bromide (CTAB) protocol following mechanical homogenization of the LMO, different crop types may require additional steps for optimal DNA extraction. It is also important to note that the pre-analytical sampling regime employed and the amount of laboratory sample used will also impact on the accuracy and sensitivity of LMO detection.
Following the extraction of DNA from a sample, target sequences only found in the LMO are amplified using primers that have been designed to specifically bind the target sequence during the PCR reaction. The resulting PCR product can either be detected in real-time during the amplification process or after the PCR is completed.
Detection for the presence of the target sequence post PCR, also known as end-point PCR, is done through the process of gel electrophoresis where the amplified target sequence is separated based on its size through a gel matrix under the influence of an electric current. The fragment of amplified DNA corresponding to the inserted DNA in the LMO can be visualised through colour detection using a dye that binds to double stranded DNA and fluoresces under ultraviolet light.
Real-time PCR technology allows the detection of the amplified target sequence during the PCR amplification process using either a fluorescent DNA binding dye or fluorescence tagged probe. The DNA binding dye simply detects the level of PCR amplification but does not discriminate between specific and non-specific amplification. In contrast, the use of a fluorescent probe can be used to verify that the specific target sequence was amplified during the PCR process. If real-time PCR technology is used in conjunction with the necessary standards, the percentage content of an LMO event in a sample can be determined. The overall applicability of this method must be evaluated in terms of the availability of equipment, its practicability and cost efficiency.
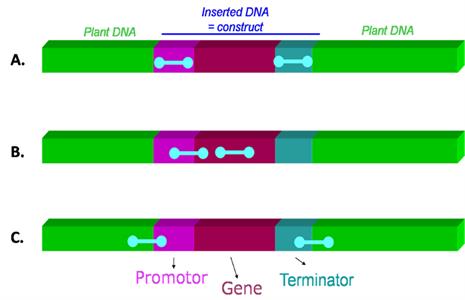
PCR has the advantage that it can be used to screen a sample for the presence LMOs by using primers that target sequences that are commonly found in a variety of different LMOs. Depending on the combination of primers used, the PCR detection can be gene-specific, construct-specific or event-specific. The differences between each type of PCR target region is represented in Figure 1. The advantage of PCR technology is that it is versatile and can be used to simultaneously screen a sample for LMO content, identify the specific LMO gene or event present and, when used in a real-time PCR platform, quantify the amount of LMO present in the sample. The disadvantage of a PCR based approach is that it requires specialized expertise and equipment.
| |
 |
| |
| Figure 1 : Different types of PCR target regions A) common regulatory elements (such as promoters, terminators) B) gene-specific or construct-specific (junction between two genetic elements within the construct) C) event-specific (junction between the inserted construct and the plant genome) Source |
|---|
2.3 Comparison between protein- and DNA-based methods for LMO detection
Both protein- and DNA-based approaches to LMO detection are useful and each serves different purposes. Protein-based methods, such as strip testing and ELISA, are simple, time efficient (several minutes to a few hours). Strip testing is useful for LMO testing at the point of sampling. However, protein detection of LMOs requires a different test for the detection of individual LMO traits and cannot be used to distinguish between different LMO events that may be present in a single sample. In contrast, western blots and PCR approaches require more steps and time (several hours to days) and rely on specialized laboratory equipment and expertise.
Protein-based methods are only suitable for detecting LMOs developed by inserting DNA fragments that produce a protein, i.e. transgenes, but not for detecting inserted genetic elements that do not produce a protein, such as regulatory sequences. Protein-based methods rely on the specific recognition of an antigen in the transgenic protein by an antibody and therefore, any changes in the tertiary structure of the protein renders the method ineffective. Such conformational changes are sometimes induced during sample processing where the samples are subjected to heat and/or chemical treatment. The detection capability in protein-based methods is also affected by the expression level of the transgenic protein that can vary between different parts of the LMO or different stages of its life cycle, and can be influenced by external factors such as climate and soil conditions. In addition, some LMOs have been specifically designed to express the transgenic protein in a specific tissue and may not be necessarily present in the part of the organism being tested.
In contrast, DNA has a greater chemical stability as compared to proteins which allows it to withstand chemical and heat treatments. DNA is also present in all cells and therefore any part of the organism can be used for testing. Furthermore, PCR methods are more versatile than protein methods and PCR can be used to screen a sample for the presence of several potential LMOs simultaneously with relative ease. In addition, PCR can be applied qualitatively, to detect specific genes or events, and quantitatively, to determine the percentage level of a particular LMO event in a sample.
3. Challenges in LMO detection and new technology developments
There are several considerations and challenges for the application of LMO detection. These include the use of validated methods, availability of reference material for specific LMO events and access to information on the LMO event including sequence data for detection method development. With the development of each new LMO, their sampling, detection and identification becomes more challenging and complex. However, progress in the field of LMO detection technologies will lead towards making LMO detection more accessible.
3.1 Method validation and quality control
An important consideration in the application of LMO detection is that standardized operating procedures (SOPs) for validated methods and equipment should be used to ensure that results are reproducible and accurate. It is therefore important that criteria to test the performance of the methods be harmonized internationally to make results comparable throughout the world taking into account the availability of resources in different countries. The development of harmonized performance criteria should aim at simplifying and increasing the accessibility to LMO detection technologies by countries with less capacity and fewer resources. Thus a challenge for LMO detection is to ensure that methods are validated and meet the necessary minimum performance criteria for quality control purposes. This means that the LMO detection laboratory is required to develop and maintain the necessary quality control measures to ensure the reliability, sensitivity and reproducibility of its methods.
3.2 Access to information and reference material for method design and validation
Another challenge in LMO detection is the difficulty for LMO detection laboratories to gain access to sequence data for genetic elements inserted into LMOs in order to design detection systems that are specific to a transformation event. While many countries require notifiers to provide a method to detect the LMO, in many cases validation of the method is not required. In addition, most regulatory LMO detection laboratories do not have access to reference material for the specific LMO event in order to validate the detection method. It may be worthwhile for regulatory systems to consider requiring LMO developers to provide reference material to the regulatory laboratory performing LMO detection for each LMO being notified to the regulatory authorities. Furthermore, since it is costly to develop and validate a new method to detect an LMO, developers could be required to provide a method for the detection of an LMO event, the necessary sequence information so that the method can be verified and to cover, as appropriate, the costs of method validation by the laboratory performing LMO detection as part of the regulatory requirements of a country.
3.3 Novel approaches for simultaneous detection of multiple LMOs
The increase in LMO research and commercialization is resulting in a continuous increase in the number of LMO events that detection laboratories must detect. As a result, the potential presence of unapproved LMO events is also increasing and detection laboratories are faced with having to potentially detect multiple LMOs, some of which may be approved while others may be unauthorised or illegal within the same sample. One approach to this challenge is to follow a matrix approach in detecting multiple genetic elements commonly used in the transgene construct in an attempt to widen the screening capabilities. When used in conjunction with bioinformatics software, the matrix results can be used to identify which potential LMOs are present in a sample. This approach can be customized to include as many LMOs as the laboratory is required to detect but is only applicable to PCR-based methods.
3.4 Emerging technologies for LMO development and detection methods
New and emerging technologies for developing LMOs are proving to be a challenge to the detection and identification of the LMOs. For example, new gene silencing technologies increasingly include using double stranded RNA (dsRNA) molecules to confer desirable traits in LMOs. In such cases, a transgenic protein is not produced therefore only DNA-based detection methods can be used.
On the other hand, the continued development of new technologies is making LMO detection more readily accessible, especially in countries with fewer resources. For example, digital PCR and other chip-based technologies will soon enable the routine detection of LMOs in the field with hand held devices. While many countries may consider the development of a framework for LMO detection to be a burden, the technology may also be applied to other purposes such as human, plant and animal pathology, amongst others.
A high-throughput multiplex assay for the detection of genetically modified organisms (GMO) was developed on the basis of the existing SNPlex method designed for SNP genotyping. This SNPlex assay allows the simultaneous detection of up to 48 short DNA sequences (70 bp; “signature sequences”) from taxa endogenous reference genes, from GMO constructions, screening targets, construct-specific, and event-specific targets, and finally from donor organisms. To overcome the difficulties of obtaining the Certified Reference Material (CRM) and according to the key documents of the European Union Reference Laboratory (EU-RL), a new standard reference molecule containing the construct specific of the canola event Oxy-235 (3'-junction Nitrilase/Tnos) and the canola endogenous reference gene (acety-CoA-carboxylase) was constructed and used for duplex real-time quantitative analysis. Description of a new strategy for the development of a
plant reference gene system that can be used for genetically modified organism (GMO) analysis. Review of new and emerging techniques in the development of analytical methods for GMO screening and quantification. A new qualitative and quantitative method based on real time polymerase chain reaction (PCR) techniques was developed for the detection and quantification of CaMV. Developed a strategy to identify unauthorised GMOs containing a pCAMBIA family vector, frequently present in transgenic plants. T This review
summarises the status of the most widely used GMO
analysis technologies, identifies new areas of analytical
investigation and discusses current needs and future
challenges. Methodological aspects in testing genetically modified plants (in the Romanian language) This "Compendium on Reference Methods for GMO Analysis" aims at providing a technical state of the art of the detection methods applied in GMO analysis that have been validated according to international standards. Discusses current detection and identification methodologies as well as highlights of new diagnostic methodologies. The aim of this report is to uncover and overview existing methods for sampling and analysing GMOs. The paper emphasizes on the need of implementing the Biosafety Protocol for the developing countries and suggests to establish capacity building measures. It identifies various ways to detect GMOs and discuss their relevance in the context of developing countries. The paper has elaborated the concept of transgene and provides an overview of GMO regulations across various countries. Based on simultaneous 24 multiplex RTi-PCR running on a ready-to-use 384-well plate, this new procedure allows the detection and identification of 47 targets on seven samples in duplicate. Herein we describe the development and validation of a pentaplex, as well as complementary triplex and duplex real-time PCR assays, for the detection of the most common screening elements found in commercialized GMOs: P-35S, T-nos, ctp2-cp4-epsps, bar, and pat. Primers and probes were developed for the
element-specific detection of cry1A.105 and cry2Ab2
genes, based on their DNA sequence as present in GM
maize MON89034 In the present study, new real-time PCR screening assays were developed targeting 10 promoter and terminator elements used in genetically modified constructs: pFMV, pNOS, pSSuAra, pTa29, pUbi, pRice actin, t35S, tE9, tOCS, and tg7. Specifically discusses methodologies that are geared towards the development of a testing system that is based on qPCR to detect and identify LMOs. This study designs four highly specific TaqMan-MGB probes. A duplex real time PCR assay was developed for simultaneous quantification of tomato and potato. For eggplant and pepper, only simplex real time PCR tests were developed. In this paper, we propose a method for the simultaneous detection
of four transgenic maize (MON810, Bt11, Bt 176, and GA21) and one transgenic soybean (Roundup Ready), which allows routine control analyses to be sped up. Describing a newly developed quantitative real-time PCR method for the detection and quantification of a new specific endogenous reference gene used in GMO analysis In this study, four novel SYBR Green qPCR methods that perform at equal efficiency were developed to allow the detection of the most common GM traits present in genetically modified crops to date ISO 21569:2005 describes the procedure to qualitatively detect genetically modified organisms (GMOs) and derived products by analysing the nucleic acids extracted from the sample under study. The main focus is on polymerase chain reaction (PCR) based amplification methods. ISO 21570:2005 provides the overall framework of quantitative methods for the detection of genetically modified organisms (GMO) in foodstuffs, using the polymerase chain reaction (PCR). Methodical Guidelines for detection and identification developed and approved in the Russian Federation and Belarus (in the Russian Language) Methodical Guidelines for detection and identification developed and approved in the Russian Federation and Belarus (in the Russian Language) This paper reviews aspects relevant to detection
and quantification of genetically modified (GM) material
within the feed/food chain. The GM crop regulatory
framework at the international level is evaluated with
reference to traceability and labelling. Current analytical
methods for the detection, identification, and quantification
of transgenic DNA in food and feed are reviewed. This paper describes a practical way to get the least expensive acceptance sampling plan keeping both the consumer’s and the producer’s risks below a
predetermined threshold. The method is more specially illustrated by examples in GMO detection. The development of four duplex polymerase chain reaction (PCR) tests for the identification and the quantification of four maize transformation events from which commercial lines have been authorised in Europe namely, Bt11 and Bt176 (Syngenta, DE, USA), Mon810 MaisGard™ (Monsanto, MO, USA) and T25 Liberty Link™ (Bayer CropScience, Monheim, Germany). The present study aims to check the status of GMO in Tunisian market using
qualitative and quantitative real time-PCR (QRT-PCR). This paper reviews the latest progress made in GMO analysis, taking examples from the most recently developed strategies and tools, and addresses some of the critical aspects related to these approaches. Presentation on the detection of unapproved GMOs from the Second International Workshop on Harmonisation of GMO Detection and Analysis, 7-8 February 2012, Nelspurit, South Africa Touches upon the isolation of DNA from processed food samples for the purposes of PCR analysis and detection as well as suggestions for circumventing potential sources of co-extracted PCR inhibitors. For the present study, the application of an informative detailed 24-element screening and subsequent identification strategy was applied in 50 animal feed samples. Almost all feed samples were labeled as containing GMO-derived materials. The main goal of the study was therefore to investigate if a detailed screening strategy would reduce the number of subsequent identification analyses. Discusses some steps in the production of reference materials. This information may be useful for those colleagues who are involved in in-house production of RM of, for example, regionally specific LMOs. Qualitative and quantitative analytical methods were developed for the new event of genetically
modified (GM) maize, MON863. Real-time PCR method for the detection of genetically
modified oilseed rape lines with the bar/T-g7-gene construct Real-time PCR assay for the quantitation of genetically modified oilseed rape lines with the 35S/pat-gene construct Critical points that affect the expression of the GMO content in seeds are discussed in this paper. A presentation on sampling methodologies from the Central and Eastern European Regional Training of Trainers' Workshop on the Identification
and Documentation of Living Modified Organisms under the Cartagena Protocol on Biosafety“, Ljubljana, Slovenia, 11-15 April 2011 This paper discusses factors that should be considered when designing and implementing seed purity testing procedures to manage this misclassification risk – especially with regard to the presence or absence of transgenic traits. A collection of statistical programs and modules to assist ISTA members in their routine work and method development and validation. In this paper, we present the details of procedures specific to quantitative laboratory methods. The scope of the training courses is to assist staff of control laboratories to become accustomed with molecular detection techniques, and to help them adapt their facilities and work programmes to include analyses that comply with worldwide regulatory acts in the field of biotechnology. The courses are intended to teach molecular detection techniques to laboratory personnel with a good level of analytical knowledge, but with no or little expertise in this specific domain. In 2008, FAO approved a two-year Technical Cooperation Programme (TCP) project in the Near East and North Africa region entitled "Strengthening capacities towards the establishment of a regional platform for the detection of genetically modified organisms", with Jordan, Lebanon, the Sudan, Syria, United Arab Emirates and Yemen as the six participating countries. As part of this TCP project, an advanced training course on "Detection of genetically modified organisms and biosafety for food and agriculture” took place in Aleppo, Syria on 19-24 June 2010, jointly organized by FAO, the International Center for Agricultural Research in the Dry Areas (ICARDA) and the General Commission for Scientific and Agricultural Research (GCSAR). In the context of this training course, a laboratory manual on GMO detection was prepared, edited by A.M. Abdul Kader et al, which is now available on the web. In this work the intra and inter-laboratory validation of a duplex real-time PCR screening method for the detection of genetically modified (gm) plants is described. Here we describe the development and validation of a
hexaplex real-time polymerase chain reaction (PCR)
screening assay covering more than 100 approved GMOs
containing at least one of the GMO targets of the assay
|